Diseases like zika and ebola can be detected using DNA, but only by testing it in hospitals or sophisticated labs. Until now.
DNA analysis is done using the Polymerase Chain Reaction (PCR) process, which creates identical copies of DNA for testing. This DNA “photocopying” became possible in 1983 thanks to Nobel Prize winner, Kary Mullis, so diseases could be identified before symptoms even arose. The process, while game-changing for molecular biology and healthcare, is extremely finicky. It must be done in a highly calibrated and strictly regulated laboratory due to the sensitivity of the PCR process and the PCR machine itself.
Biomedical engineers at Vanderbilt University recently enhanced the PCR process, which will drastically shrink the size of the PCR machine to hand-held. This means the analysis could be used in the field, far away from hospitals and sophisticated labs.
The PCR process is sensitive and complex.
Chemical composition and temperature are key. Any minor variations in the samples’ chemistry or the room in which the massive PCR machine is can affect the results. This is why extensive laboratory calibration is required and must be maintained for the process to be successful.
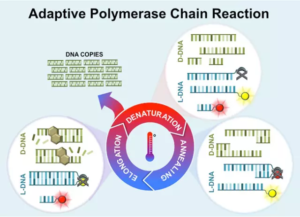
The improved process is called “adaptive PCR”, which uses synthetic, commercially-available left-hand DNA (L-DNA) to monitor and control the reactions in the PCR process. L-DNA doesn’t replicate but is otherwise identical to right-handed DNA and is thus a controlled variable in the process.
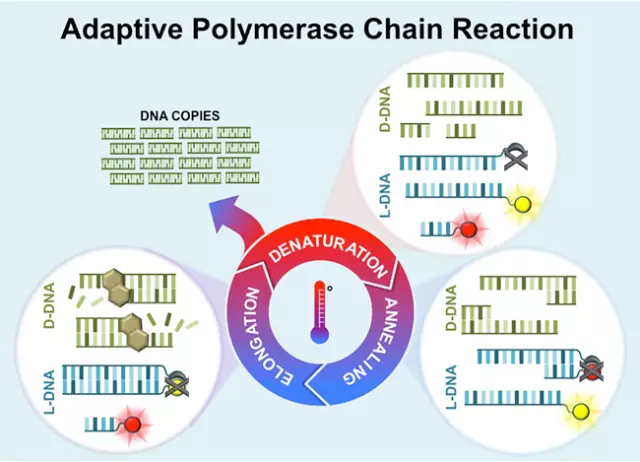
When adding fluorescently tagged L-DNA to the PCR sample, it not only provides valuable information about the PCR reactions but can also control them. This means that the required conditions for the process are determined by the L-DNA, not laboratory technicians. The L-DNA literally lights up while the DNA sample is being processed, providing technicians with valuable information. Samples can also be tested for multiple diseases at the same time using this technique.
With this new approach, the PCR process can be performed using a hand-held device at a patients’ bedside anywhere in the world. It drastically reduces the sensitivity of the process, while increasing its effectiveness and efficiency. The adaptive PCR process could also be used for other applications, including food safety and bioterrorism.
The Vanderbilt engineers expect it to be commercially available in the next few years.
Source: http://www.engineering.com/


















